IPython Notebook
Recently, I've been using IPython notebook for some data analysis. It's pretty janky in places, like most browser-based software, but it's the closest thing Python has to an interactive environment. It definitely saves a bunch of time if you have a long series of independent data transformations and analyses.
Ideally, I'd be able to work on a notebook locally, but if I have some heavy computation to do, I could transparently send that to the cloud. With IPython it's technically possible to do that on a cell by cell basis using ipcluster, but it would be difficult to integrate that with a third-party cloud provider. It's also not an elegant system: for example, you have to manually do imports on each of your ipcluster nodes.
A simpler method is just to dump the IPython notebook to a Python file, then send the entire script to the cloud. Assuming there is one long-running bottleneck task in there, this should take about the same amount of time to run.
Using Domino/AWS with IPython
I'm still not convinced about Domino as an AWS intermediary — for example, once my free trial ends I think am limited to just one project on there — but that's ok since the following process has only a few Domino-specific elements.
requirements.txt
I needed to add seaborn since it was not included in Domino's standard set of imports. For some reason I needed to include numexpr to get pandas to work. It's probably a good idea to include as little as possible in requirements.txt (i.e., don't pip freeze) since Domino has to install everything you ask for anew with each run.
seaborn==0.3.1 numexpr==2.3.1
IPython setup
It's important that I use all available CPUs when I am running on AWS/Domino, otherwise I am wasting money. I need a few CPUs free on my laptop though... I test this by just checking if the platform is Linux.
import platform, multiprocessing N_CPUS = multiprocessing.cpu_count() if platform.system() == 'Linux' else 5
IPython's nbconvert preprocessor
IPython's handy nbconvert function converts IPython notebooks into other formats: most commonly, pure Python, HTML or PDF.
An IPython preprocessor is a little function that takes the cells in your notebook, and does something to them before nbconvert gets to them. Here I am using a preprocessor to do a few little things:
- comment out IPython "magic" commands (these will cause errors if run outside IPython)
- skip cells that have a special "SKIPCELL" comment (surprisingly, there's no IPython magic command for this)
- make the "print" function print to a file. For some reason Domino appears to munge stdout and stderr into one file. By making the "print" function print to a file I can separate my results from warnings etc.
I'd also like to dump all my inline plots to files, but I don't know how to do that.
c = get_config() #Export all the notebooks in the current directory to the sphinx_howto format. c.NbConvertApp.notebooks = ['kaggle.ipynb'] c.NbConvertApp.export_format = 'python' c.Exporter.preprocessors = ['domino_preprocessor.DominoPreprocessor']
from nbconvert.preprocessors import * class DominoPreprocessor(Preprocessor): FIRSTCELL = None print_fn = """ global_out = open("stdout.ipy.txt",'w') def print(*args, **kwargs): kwargs['file'] = global_out return __builtins__.print(*args, **kwargs) """ def preprocess_cell(self, cell, resources, index): if cell.cell_type == 'code': SKIPCELL = True if "# SKIPCELL" in cell.source else False DominoPreprocessor.FIRSTCELL = True if DominoPreprocessor.FIRSTCELL is None else False newlines = [] for line in cell.source.splitlines(): if line.startswith('%'): line = "## "+line elif SKIPCELL: line = "# "+line newlines.append(line) if DominoPreprocessor.FIRSTCELL is True: newlines.append(DominoPreprocessor.print_fn) DominoPreprocessor.FIRSTCELL = False cell.source = '\n'.join(newlines) return cell, resources
Running the code on Domino
Finally, I created a small script that generates the cleaned-up Python file, sends it to Domino to run and opens the results page in a browser window.
#!/usr/bin/env python # # Create domino-appropriate python file # from subprocess import Popen, PIPE p = Popen(["ipython", "nbconvert", "--config", "domino_config.py"], stdin=PIPE, stdout=PIPE, stderr=PIPE) out1, err1 = p.communicate() # # Upload to Domino # p = Popen(["/Applications/domino/domino", "run", "kaggle.py"], stdin=PIPE, stdout=PIPE, stderr=PIPE) out2, err2 = p.communicate() # # Show the results page # import re, webbrowser rc = re.compile('(https://app.dominoup.com/[\S]+)') url = rc.search(out2).group(1) webbrowser.open(url)
Using this script I can trivially run my Kaggle script on 32 CPUs without sending my laptop into paroxysms. All in all it works pretty well.
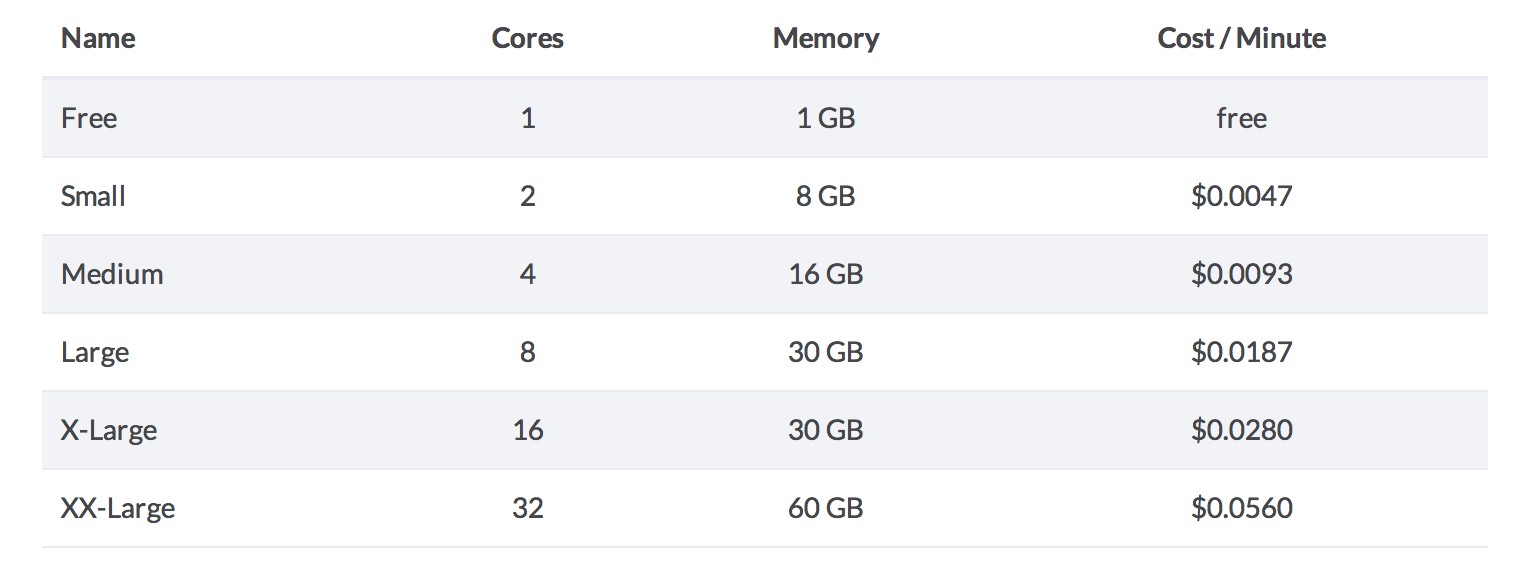
Domino Pricing vs AWS
Domino sells standard "Compute Optimized" AWS instances, and marks them up 100%. It's not a bad deal for light to moderate users, considering that the AWS environments are already spun up and waiting, and that Domino charges by the minute instead of by the hour. For comparison purposes, 32 CPUs costs $1.68 an hour on AWS vs $3.36 on Domino.